Multivariate Statistical Process Control with mvMonitoring
Gabriel Odom
2023-11-21
Source:vignettes/MVSPC-Workflow.Rmd
MVSPC-Workflow.RmdThis document serves to introduce engineers to the workflow necessary
to follow in order to analyze multivariate process monitoring data via
the mvMonitoring package.
Introduction
This is the accompanying package to the research published by Kazor et al (2016)
and Odom et al (2018).
The mvMonitoring package is designed to make simulation of
multi-state multivariate process monitoring statistics easy and
straightforward, as well as streamlining the online process monitoring
component.
Motivation
Why use mvMonitoring
The mvMonitoring package can be used to detect outliers
in a correlated multivariate process with non-linear, non-stationary, or
autocorrelated feature behavior. These process outliers are often
indicative of system fault. The common and naive approach to
multivariate process monitoring is to use expert opinion to identify a
few important features to monitor visually, and raise an alarm if these
features travel outside pre-defined normal operating boundaries.
However, this split univariate approach fails to account for the correlated nature of the process features, so some engineers have taken to monitoring the system as a single correlated multivariate process rather than a collection of independent univariate processes. In the literature review of the motivating papers, the authors cite how this approach has benefitted the science of process monitoring as a whole. Unfortunately, this approach has its own shortcomings – namely, monitoring a multivariate process in its original feature space can lead to exorbitant computational costs.
To this end, principal components analysis (PCA) and its many modifications have been employed to reduce the computation necessary to monitor a multivariate process as a whole. This package is an implementation of one such PCA modification.
Multi- or Single-State AD-PCA
AD-PCA: adaptive-dynamic PCA, which accounts for non-linearity, non-stationarity, and autocorrelation in the multivariate process. This modification is thoroughly discussed in Kazor et al (2016).
Multi-State Monitoring: a modification of PCA which accounts for multiple process states and models them separately. States can be any mutually exclusive blocking factor, and states do not necessarily follow a strict order.
Multi-State AD-PCA (MSAD-PCA) allows process engineers to account for a few very different process states. Specifically, this modification should be used when features under different process states have different means, correlations, or variances.
The msp Functions
These are brief descriptions of the external functions useful for multivariate statistical process monitoring via MSAD- or AD-PCA.
Simulating Data with mspProcessData
This function generates three-dimensional multi-state or single-state non-linear, non-stationary, and autocorrelated process observations. We follow the seminal work of Dong and McAvoy (1996) for generation of the foundational stationary and independent features.
Latent Feature Creation
So that the simulated features have non-zero correlations, Dong and McAvoy created their three features all as polynomial functions of a single latent variable \(t(s)\), where \(s = 1, \ldots, \omega\) is the observational index of the process.
Autocorrelated and Non-Stationary Error
The mspProcesData function induces autocorrelation in
\(t\) through its errors, \(\varepsilon_s\), where \[
\varepsilon_1 \sim \mathcal{N}\left(\frac{1}{2}(a + b)(1 - \phi),
\frac{b - a}{12} (1 - \phi ^ 2)\right),
\] where \(a = 0.01\) and \(b = 2\). Now, we define the first-order
autoregressive process on \(\varepsilon_s\) by \[
\varepsilon_s = \varphi\varepsilon_{s - 1} + (1 - \varphi)\varepsilon,
\] where \(\varepsilon\) is as
defined in the previous expression and the autocorrelation component
\(\phi = 0.75\). The mean and variance
multipliers are the mean and variance of a random variable from the
\(\mathcal{U}_{[a,b]}\)
distribution.
The Non-Linear Latent Process
This \(t\) vector will be sinusoidal with period \(\omega = 7 * 24 * 60\) (signifying a weekly period in minute-level observations). We then synthesize a \(t\) with \[ t^*_s = -\cos\left( \frac{2\pi}{\omega} s \right) + \epsilon_s, \] and scale \(t^*\) to \[ t = \frac{(b - a)(t^*_s - \text{min}(t^*_s))}{\text{max}(t^*_s) - \text{min}(t^*_s)} + a. \] Finally then, the \(t\) vector will lie entirely in \([a,b]\).
Single-State and Multi-State Features
Single State Features
First mspProcessData simulates three features, with each
feature operating under \(k\) different
states. Let \(<x_k(t), y_k(t),
z_k(t)>\) be the notation for State \(k\). These are the three features under
State 1 (normal operating conditions, or NOC) as three
functions of \(t\): \[\begin{align}
\textbf{x}(\textbf{t}) &\equiv \textbf{t} +
\boldsymbol\varepsilon_1, \\
\textbf{y}(\textbf{t}) &\equiv \textbf{t} ^ 2 - 3 * \textbf{t} +
\boldsymbol\varepsilon_2, \\
\textbf{z}(\textbf{t}) &\equiv -\textbf{t} ^ 3 + 3 * \textbf{t} ^
2 + \boldsymbol\varepsilon_3,
\end{align}\] where \(\varepsilon_i
\sim N(0, 0.01)\). The mspProcessData function calls
the internal processNOCdata function to generate
single-state observations under normal operating conditions (NOC).
Multi-State Features
The multi-state feature expression is induced by rotation and scaling
of certain sets of observations. To induce a three-state, hourly
switching process (the default), the mspProcessData
function will create a label column that switches from “1” to “2” to “3”
every hour. State “1” will be the features generated under the
single-state assumption, while State “2” and State “3” are generated as
follows. These states will be scaled rotations of the current \(<x,y,z>\) set. The second state is
yaw, pitch, and roll rotated by (0, 90, 30) degrees, and the scales are
multiplied by (1, 0.5, 2). The third state is yaw, pitch, and roll
rotated by (90, 0, -30) degrees, and the scales are multiplied by (0.25,
0.1, 0.75).
\(\mathcal{S}_1\): \(\textbf{X}(t_s) := \langle x(t_s), y(t_s), z(t_s)\rangle\).
\(\mathcal{S}_2\): \(\textbf{X}(t_s) := \langle x(t_s), y(t_s), z(t_s)\rangle \cdot \textbf{P}_1\boldsymbol\Lambda_1\), where \[ \textbf{P}_1 = \begin{bmatrix} 0 & 0.50 & -0.87 \\ 0 & 0.87 & 0.50 \\ 1 & 0 & 0 \end{bmatrix} \] is the orthogonal rotation matrix for a yaw, pitch and roll degree change of \(\langle 0^{\circ}, 90^{\circ}, 30^{\circ} \rangle\), and \(\boldsymbol\Lambda_1 = \text{diag}(1, 0.5, 2)\) is a diagonal scaling matrix.
\(\mathcal{S}_3\): \(\textbf{X}(t_s) := \langle x(t_s), y(t_s), z(t_s)\rangle \cdot \textbf{P}_2\boldsymbol\Lambda_2\), where \[ \textbf{P}_2 = \begin{bmatrix} 0 & 0.87 & -0.50 \\ -1 & 0 & 0 \\ 0 & 0.50 & 0.87 \end{bmatrix} \] is the orthogonal rotation matrix for a yaw, pitch and roll degree change of \(\langle 90^{\circ}, 0^{\circ}, -30^{\circ} \rangle\), and \(\boldsymbol\Lambda_2 = \text{diag}(0.25, 0.1, 0.75)\) is a diagonal scaling matrix.
These rotation matrices \(\textbf{P}_1\) and \(\textbf{P}_2\) turn the states in
three-dimensional space so that the states are at right angles to each
other in at least one dimension, and the scaling matrices \(\boldsymbol\Lambda_1\) and \(\boldsymbol\Lambda_2\) inflate or deflate
the process variances along each principal component. The
mspProcessData function calls the internal function
dataStateSwitch which splits the observations by state and
applies the state-specific rotation and scaling through the internal
rotateScale3D function.
Synthetic Fault Induction
Faults can be introduced to single- or multi-state data via the
mspProcessData function. The default fault start index is
8500, or roughly 84% through the 10,080 observation cycle. These faults
are added through the internal faultSwitch function.
Fault 1A is a positive shift to all three features before state rotation: \(\textbf{X}^*(t_s) = \textbf{X}(t_s) + 2,\ s \ge 8500\).
Fault 1B is a positive shift to the \(x\) feature before state rotation: \(x^*(t_s) = x(t_s) + 2,\ s \ge 8500\).
Fault 1C is a positive shift to the \(x\) and \(z\) features in State 3 only and after state rotation: \(x^*(t_s) = x(t_s) + 2,\ z^*(t_s) = z(t_s) + 2,\ s \ge 8500\).
Fault 2A is a positive drift across all the process monitoring features before state rotation: \(\textbf{X}^*(t_s) = \textbf{X}(t_s) + (s - 8500)\times 10^{-3}\), \(s > 8500\).
Fault 2B is a positive drift across the \(y\) and \(z\) process monitoring features before state rotation: \(y^*(t_s) = y(t_s) + (s - 8500)\times 10^{-3},\ z^*(t_s) = z(t_s) + (s - 8500)\times 10^{-3}\), \(s > 8500\).
Fault 2C is a negative drift in the \(y\) process monitoring feature in State 2 only and after state rotation: \(y^*(t_s) = y(t_s) - 1.5 \times \frac{s - 8500}{10080 - 8500}\), for \(s > 8500\).
Fault 3A is an amplification of the underlying latent variable \(t\) for all features. The maximum latent drift of this fault will be 5 + 1: \(\textbf{X}^*(t_s) = \textbf{X}(t_s^*),\ s > 8500\), where \(t_s^* = \left[\frac{5(s - 8500)}{\omega - 8500} + 1\right]t_s\).
Fault 3B is a mutation of the underlying latent variable \(t\) for the \(z\) feature: \(z^*(t_s) = z(\log t_s^*)\), \(s \ge 8500\). This fault will dampen the underlying latent effect for \(z\) if \(t_s > 1\) and amplify this effect if \(t_s < 1\).
Fault 3C is a polynomial mutation of the error for the \(y\) feature in State 2 only and after state rotation: \(y^*(t_s) = y(t_s) + 2 * \textbf{e}_3(s) - 0.25\), for \(s > 8500\).
Putting it all together…
The mspProcessData function can generate weeks’ worth of
non-linear, non-stationary, autocorrelated, multi-state, multivariate
process data useful to test new process monitoring techniques. Users can
generate observations under NOC to measure false alarm rates, or induce
one of nine pre-built faults to test detection time and consistency with
repeated Monte Carlo sampling. We expect this function will generate
interesting data useful enough to compare new and improved process
monitoring techniques with existing methods.
Training with mspTrain
The mspTrain function will generate projection matrices
and test statistic thresholds from test data matrices.
The xts Data Matrix
An xts data matrix is first and foremost a matrix,
not a data frame. For users very familiar with data frame
manipulation (with dplyr for instance), the slight but
profound differences between manipulating matrices and data frames
become quickly apparent. Because of the class requirements for matrices,
all features must be integer or double objects. The
mspTrain function cannot train on character
information.
The xts object class stands for extendable time
series and comes from the package xts, which is itself
built on the package zoo. The date and time information
(necessarily as POSIX objects) are stored as the row
indices of xts matrices. We recommend the package
lubridate for manipulating POSIX objects.
The Class Vector
When implementing single-state AD-PCA, this class vector simply needs
to be a numeric column of the same value. However, for MSAD-PCA, the
class vector should be an integer vector of class membership. The
mspTrain function will split the observations by the class
label, apply single-state AD-PCA to each class, then return the
class-specific projection matrices and thresholds. Because of this
split-apply-combine strategy, users must ensure that one or more classes
are not too “rare” – that is, the class-specific sample size should be
sufficiently large to allow for stable covariance matrix inversion. For
\(p\) features (including lags),
covariance inversion requires a class sample size greater than \(p ^ 2 / 2\). Because of this, pay close
attention to model parsimony – don’t block the observations on a factor
unless the factor significantly affects the observations’ mean vector or
covariance matrix.
Adaptive and Dynamic Modelling
Because of the non-linear, non-stationary, and autocorrelated nature
of some process monitoring applications, the mspTrain
function allows users to include lags of all feature variables and also
to update the training window over time. Including lags of the features
in the data matrix can significantly reduce the negative effects of
autocorrelation. Re-estimating the projection matrix and test statistic
thresholds at pre-specified time intervals reduces the negative effects
of non-linearity and non-stationarity in the observations.
The idea is to divide a non-linear and non-stationary process along some boundaries (every day, for instance), and then the process becomes locally linear and stationary within the window. As time progresses, the oldest observations are “forgotten” and the newest observations are “learned”, which causes the projection and NOC thresholds to update over time.
Model Training
After the observations have been split by class, the
mspTrain function will call the internal function
processMonitor, which in turn calls the internal function
faultFilter. This function will calculate a linear
projection matrix of the data by taking the PCA of the training data
matrix. The observations will then be projected linearly into a
reduced-feature subspace which preserves a chosen proportion of the
energy of the training data, where the energy of a matrix is the sum of
eigenvalues of that matrix. The default proportion is 90%. This
projection is calculated by the internal pca function.
Furthermore, non-parametric threshold values are calculated for the
two process monitoring statistics – Squared Prediction Error
and Hotelling’s \(T^2\). These
monitoring statistics are described in the motivating paper. These
threshold values are found in the internal threshold
function, and passed through the function pipe to be returned by
mspTrain. The \(\alpha\)-level of the nonparametric
threshold is controlled by the user, and its default value is 0.001.
Finally, the mspTrain function will remove any
observation which would cause an alarm from the training data set. The
alarm free observations will be returned in one xts matrix,
while the alarmed observations will be returned by another. When
training your process monitor, pay attention to the observations flagged
as alarms. The proportion of observations flagged as faults may be
higher than the \(\alpha\)-level
specified, so some tuning may be necessary.
Monitoring Incoming Observations with mspMonitor
After training the model with mspTrain, the projection
matrices and non-parametric monitoring statistic thresholds can be used
to flag incoming observations. This function can check every observation
in a test matrix (useful when analyzing past data), but was instead
designed to test a single incoming observation at a time via a script or
batch file. To this end, the mspMonitor function projects a
single observation with the class projection matrices returned by
mspTrain and checks the observation’s SPE and \(T^2\) statistics against the thresholds
returned by mspTrain. The mspMonitor will then
append the monitoring statistic values, and indicators if these values
exceed normal thresholds, to the end of the observation row. This new
appended observation will be passed to the mspWarning
function.
Issue a Warning with mspWarning
If an observation returned by the mspMonitor function
has positive statistic indicator values for either the SPE or \(T^2\) monitoring statistics, then the
mspWarning function will query the last set \(r\) observations for other flags. If all
\(r\) observations are positive for
anomalies, then the mspWarning function will issue an
alarm. This is the parameter “faultsToTriggerAlarm”, and it defaults to
5. However, this default value depends heavily on the scale of the data:
for continuous observations aggregated and recorded every five seconds,
the number of sequential faults necessary to trigger an alarm should be
very high, perhaps even 50 or more. For observations aggregated to the
10-minute-scale, only three sequential faults may be necessary.
In future updates of this package, this function will also have an option to issue an alarm if a critical mass of non-sequential alarms is reached in a set period of observations. This modification may be necessary if the observation level becomes more fine than the 1-minute-level. Additionally, this function will also be equipped to take in a cell phone number and service provider and issue an alarm via SMS through email.
Example Simulation Workflow
This section provides a fully-commented code walkthrough for the main
msp functions in the mvMonitoring package.
Generating Synthetic Data
First begin by generating multi-state data from a fault scenario.
This code will yield observations under Fault 2A, as described the
Synthetic Fault Induction section. We choose the default options for the
period length (7 days * 24 hours * 60 minutes = 10,080 observations),
the starting index of the fault (8500 out of 10080), and the time stamp
for beginning the data is 16 May of 2016 at 10:00AM (my wedding
anniversary). As we can see from the str() function, we
have an xts matrix with 10080 rows and four columns (the
state indicator and the three features).
library(mvMonitoring)
fault1A_xts <- mspProcessData(faults = "A1",
period = 7 * 24 * 60,
faultStartIndex = 8500,
startTime = "2015-05-16 10:00:00 CST")
str(fault1A_xts)## An xts object on 2015-05-16 10:00:00 / 2015-05-23 09:59:00 containing:
## Data: double [10080, 4]
## Columns: state, x, y, z
## Index: POSIXct,POSIXt [10080] (TZ: "")Train the Fault Detection Threshold
Now that these observations are generated and stored in memory, the
mspTrain function can train the MSAD-PCA model. The last
1620 observations (27 hours’ worth) will be saved for testing. The
mspTrain function takes in the training data set
partitioned between the observation and the label column. If this
function errors, make sure the label column is not included in the data
matrix – this will cause a singularity in the data. The function will 1.
Train on the first three days’ worth of observations, as set by
trainObs. 1. Scan the fourth day for anomalies, as set by
updateFreq. 1. Remove any alarmed observations. 1. “Forget”
the first day’s observations. 1. “Learn” the non-alarmed observations
from the fourth day. 1. Retrain and repeat until the end of the data
matrix
Furthermore, the Dynamic = TRUE option means that the
mspTrain function will include the lags specified by the
lagsIncluded argument. Finally, the number of sequential
anomalous observations necessary to raise an alarm is set at 5 by the
faultsToTriggerAlarm argument. These last three arguments
are set to their defaults.
train1A_xts <- fault1A_xts[1:8461,]
# This function will run in 13 seconds on the author's machine.
train1A_ls <- mspTrain(data = train1A_xts[,-1],
labelVector = train1A_xts[,1],
trainObs = 3 * 24 * 60,
updateFreq = 1 * 24 * 60,
Dynamic = TRUE,
lagsIncluded = 0:1,
faultsToTriggerAlarm = 5)## Warning: In density.default(spe, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(t2, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(spe, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(t2, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(spe, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(t2, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(spe, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(t2, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(spe, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(t2, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(spe, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(t2, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(spe, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(t2, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(spe, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(t2, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(spe, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregarded## Warning: In density.default(t2, bw = "SJ", kernel = "gaussian", from = 0,
## old.coords = TRUE) :
## extra argument 'old.coords' will be disregardedThe mspTrain function returns a list of four
objects:
-
FaultChecks: Anxtsmatrix of monitoring statistics and associated indicators for all observations after the burn-in oftrainObs. It will have 10080 -trainObsnumber of rows and five columns:-
SPE: the SPE statistic for each observation. -
SPE_Flag: an indicator showing if the SPE statistic for that observation is beyond the calculated threshold; 0 is normal, 1 is flagged. -
T2: the \(T^2\) statistic for each observation. -
T2_Flag: an indicator showing if the \(T^2\) statistic for that observation is beyond the calculated threshold; 0 is normal, 1 is flagged. -
Alarm: an indicator showing if the observation is in a sequence of flagged observations; 0 is normal, 1 is alarmed.
-
-
Non_Alarmed_Obs: Anxtsmatrix will all observations with an alarm code of 0 fromFaultChecks. Of note, this matrix contains the data, while theFaultChecksmatrix only contains the monitoring statistics and indicators. -
Alarms: Anxtsmatrix of all the observations removed from the training data matrix. -
TrainingSpecs: a list with length equal to the number of classes – in this case 3. For each class, this list contains a list of six objects:-
SPE_Threshold: a named numeric scalar of the 1 - \(\alpha\) percentile of the non-parametric estimate of the SPE statistic density. -
T2_Threshold: a named numeric scalar of the 1 - \(\alpha\) percentile of the non-parametric estimate of the \(T^2\) statistic density. -
projectionMatrix: The \(p \times q\) matrix of eigenvectors necessary to project a \(p\)-dimensional observation to \(q\) dimensions. This is necessary to reduce the dimension of any test observation, and is used in calculating the SPE statistic for test observations. -
LambdaInv: The inverse of the diagonal \(q \times q\) matrix of eigenvalues. This matrix is used to calculate the \(T^2\) statistic for test observations. -
muTrain: The mean vector of the training observations. This is used to center the test observations on the training mean. -
RootPrecisTrain: The \(p \times p\) diagonal matrix of the inverse square roots of the feature variances. This is used to scale the test observations into the training scale.
-
Test New Observations for Anomalies
The training data summary was given by mspTrain, so this
information can now be used to monitor incoming observations for system
faults.
Adding Lagged Features
First, concatenate the last given observation from the training set
as “row 0” of the test data set. This will enable
mspMonitor to include lag-1 features. Similarly, one would
include the last \(k\) observations of
the training set should the process dictate the need for any lag-\(k\) features. Because the Fault Start Index
was set to 8500, this testing window will show the change point between
observations generated under normal conditions and those generated under
a fault state.
library(xts)
test1A_xts <- fault1A_xts[8460:8520, -1]
lagTest1A_xts <- lag.xts(test1A_xts, 0:1)
lagTest1A_xts <- cbind(fault1A_xts[8461:8520,1],
lagTest1A_xts[-1,])
head(lagTest1A_xts)## state x y z x.1 y.1
## 2015-05-22 07:00:00 1 0.5357652 -1.350724 0.7527411 0.3440182 0.0806325
## 2015-05-22 07:01:00 1 0.6470256 -1.332191 0.8228733 0.5357652 -1.3507237
## 2015-05-22 07:02:00 1 0.6169399 -1.523472 1.0138078 0.6470256 -1.3321906
## 2015-05-22 07:03:00 1 0.6096512 -1.618545 0.9675310 0.6169399 -1.5234719
## 2015-05-22 07:04:00 1 0.7532528 -1.568173 1.0334942 0.6096512 -1.6185449
## 2015-05-22 07:05:00 1 0.6001876 -1.630557 0.9020732 0.7532528 -1.5681727
## z.1
## 2015-05-22 07:00:00 0.2486086
## 2015-05-22 07:01:00 0.7527411
## 2015-05-22 07:02:00 0.8228733
## 2015-05-22 07:03:00 1.0138078
## 2015-05-22 07:04:00 0.9675310
## 2015-05-22 07:05:00 1.0334942Monitoring the Test Data
With the lagged test observations in the working environment, the
mspMonitor function can be applied. This function
(similarly to mspTrain) takes in the label information as a
separate argument from the input data. Further, the
mspMonitor function takes in the TrainingSpecs
object returned in the results list from mspTrain. Notice
that the first six rows of the monitor matrix are the exact same as the
first six rows of the lagged test matrix, except that the rows of the
monitor matrix have the monitoring statistic and corresponding indicator
columns appended.
monitor1A_xts <- mspMonitor(observations = lagTest1A_xts[,-1],
labelVector = lagTest1A_xts[,1],
trainingSummary = train1A_ls$TrainingSpecs)
head(monitor1A_xts)## x y z x.1 y.1
## 2015-05-22 07:00:00 0.5357652 -1.350724 0.7527411 0.3440182 0.0806325
## 2015-05-22 07:01:00 0.6470256 -1.332191 0.8228733 0.5357652 -1.3507237
## 2015-05-22 07:02:00 0.6169399 -1.523472 1.0138078 0.6470256 -1.3321906
## 2015-05-22 07:03:00 0.6096512 -1.618545 0.9675310 0.6169399 -1.5234719
## 2015-05-22 07:04:00 0.7532528 -1.568173 1.0334942 0.6096512 -1.6185449
## 2015-05-22 07:05:00 0.6001876 -1.630557 0.9020732 0.7532528 -1.5681727
## z.1 SPE SPE_Flag T2 T2_Flag Alarm
## 2015-05-22 07:00:00 0.2486086 0.6528551 0 26.563188 0 NA
## 2015-05-22 07:01:00 0.7527411 0.8902785 0 5.856376 0 NA
## 2015-05-22 07:02:00 0.8228733 0.1903380 0 4.952093 0 NA
## 2015-05-22 07:03:00 1.0138078 0.0181310 0 4.264220 0 NA
## 2015-05-22 07:04:00 0.9675310 0.1974976 0 4.024429 0 NA
## 2015-05-22 07:05:00 1.0334942 0.1604523 0 4.052323 0 NAWarn Operators during Alarms
Note that all Alarm codes in the monitor matrix above
are recorded with NA values. This is because the
mspMonitor function does not check the sequential flag
conditions of the monitor matrix. This is the responsibility of the
mspWarning function. Because the mspWarning
function is designed to test one incoming observation at a time through
a script or batch file, the following example is designed to mimic the
behavior of the mspWarning function as each new observation
comes online.
alarm1A_xts <- monitor1A_xts
for(i in 1:nrow(alarm1A_xts)){
if(i < (5 + 1)){
alarm1A_xts[1:i,] <- mspWarning(alarm1A_xts[1:i,])
}else{
alarm1A_xts[(i - 5):i,] <- mspWarning(alarm1A_xts[(i - 5):i,])
}
}The fault was introduced at index 8500, which corresponds to about 40 minutes into the test hour.
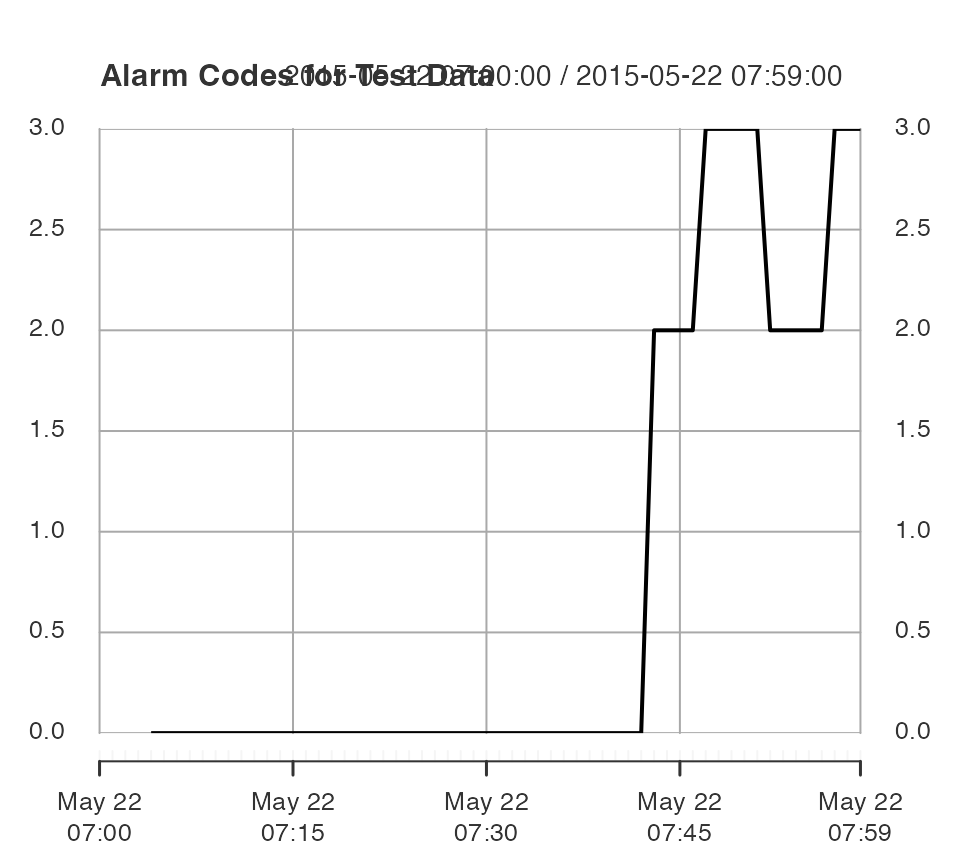 The alarm codes are
The alarm codes are
- “0”: No alarm.
- “1”: Hotelling’s \(T^2\) alarm.
- “2”: Squared Prediction Error alarm.
- “3”: Both alarms.
Conclusion
We have supplied our motivation for this package, and we have discussed implementing a multivariate process monitoring scheme with this package using the example of a decentralized WWT plant in Golden, CO. We believe that this software will provide system engineers with the tools necessary to quickly and accurately detect abnormalities in multivariate, autocorrelated, non-stationary, non-linear, and multi-state water treatment systems. Further, we have given a synthetic example showing how the functions within this package would be implemented and tuned in practice.